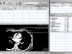
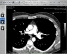
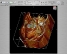
OSIRIX
OSIRIX is a free software that is available from http://homepage.mac.com/rossetantoine/osirix/Index2.html. It is basically a DICOM viewer that can do many basic things to assist in general medical visualization and analysis.
Obtaining and installing OSIRIX is trivial and i will not elaborate on it.
Some Terms:-
- WL/WW: Window Width and Window Level (Some of the presets are available for various CT scans in the drop down list).
- CLUT: Color Look Up Table for various parts of the body. Some presets are available
- MIP: Maximum Intensity Projection
- MinIP: Minimum Intensity Projection
Quick guide
Here are some basic procedures that i have tested.
- Open a CT scan. It can be a series of images.
- Use the scroll wheel to browse through the series of images
- click on BROWSE for playback of the series
- Select Mode – Volume rendering UP/DOWN to show build up or down of image series
- Select 2D/3D
- use 2D MPR to be able to choose the cut plane section
- move the blue line
- move the red circle
- able to get angle, identify region size and measure
- Right click
- ability to select tools for ROI and annotation of series
- ROI menu item is able to give the ability to manage a list of ROIs
- Select 2D/3D
- select 3D MIP (another method of rendering the data)
- set the details to FINE
- set the Engine to GPU, Raycast or CPU depending on hardware
- select CROP to be able to crop the model at any region
- Select 2D/3D
- selection 3D Volume Render
- select Jolly Roger and pick areas to remove. Can be bones or arteries
- Use GPU render for improved performance if a graphic card is present
- select the scissors tool to select region to be cut (delete button, tab to restore)
- select FLYTHROUGH and set the various key points to allow views to be composed into a movie.
- 2D Orthogonal MIP
- Selects all 3 views for a given CT scan
- Select MIP mode and adjust the slab thickness
- Scroll through the data set by clicking on the blue lines and moving it around. NOTE that the blue lines represent the thickness of the slab.
- To Compare Cases (This is great to compare a set of cases)
- Apple Select the cases that you wish to compare
- Select the Sync item from the toolbar to sync the cases at the slice point. The cases will move along the same slice point as the scroll wheel is moved.
- To propagate changes such as rotate between windows of the same view, one can use the “Propagate” item in the toolbar
- To explore the meta-data, one can use the “Meta-Data” item in the toolbar
- Right click to select tools.
- Use ROI tools to measure various components and double click (while on ROI tool) to show it on the other views
- We can KEY the image to highlight (in yellow) the image of interest
- Selection available from the right click menu
- To Annotate, use ROI->Text and double click to edit the comments on that image. One can move the comments around.
- To Fuse Image
- Open the images to use
- Drag icon on window title to the target image to fuse with
- Select Image Fuse
PACS Server with dcm4chee
In order to implement any for of DICOM viewer in a production environment, a PACS server is required to assist in storing an classifying the various information that is coming in form the CT, PT or MR scans. Though i am no expert in this area, i have come to realize that these files can be huge, easily reaching a few gigabytes for a simple scan.
Setting up dcm4chee
(Base reference http://homepage.mac.com/rossetantoine/osirix/Index2.html)
There are 2 choices
- CDMedic PACS Web
- dcm4chee
I am more interested in installing dcm4chee as it does not require FINK installation
Required Files
- dcm4chee-standalone-mysql-2.10.10.zip (http://downloads.sourceforge.net/dcm4che/dcm4chee-standalone-mysql-2.10.10.zip)
- MySQL (http://www.mysql.org/downloads/mysql/5.0.html)
Here is the installation procedure
- Install all components of mySQL sever
- unzip dcm4che package
- “cd” to the package location
- NOTE that mysql will be installed into /usr/local/mysql. The commands for mysql needs to be accessing the “mysql” binary from “/usr/local/mysql/bin”
- cd /usr/local/mysql/bin
- Run the following commands to configure the database:-
- mysql -uroot
- create database pacsdb;
- grant all on pacsdb.* to ‘pacs’@’localhost’ identified by ‘pacs’;
- quit
- To check the entries
- mysql -uroot
- use mysql;
- show grants for ‘pacs’@’localhost’;
- If a mistake was made, you can drop the user by using
- drop user ‘pacs’@’localhost’;
- ./mysql \pacsdb \-upacs \-ppacs < /Users/rossetantoine/Desktop/dcm4chee-standalone-mysql-2.10.10/sql/create.mysql
- check the mysql import
- mysql -uroot
- use pacsdb;
- show tables;
- quit;
- Add the following lines to the “/etc/bashrc” file
- # For dcm4che DICOM PACS Database
- export JAVA_HOME=”/System/Library/Frameworks/JavaVM.framework/Home”
- export JAVA_OPTS=”-server -Xms128m -Xmx200m -Djava.awt.headless=true -Djava.library.path=. -Dsun.rmi.dgc.client.gcInterval=3600000 -Dsun.rmi.dgc.server.gcInterval=3600001″
- source /etc/bashrc
- ensure that you are not root
- execute [dcm4chee-package]/bin/run.sh and watch the output
- Log into server using http://pacs-server:8080/dcm4chee-web (username and password is “admin:admin”)
Configuring OSIRIX to talk to the dcm4chee PACS server
- Start OSIRIX and select Preferences-Locations
- Add dcm4chee as a PACS server. Fields as follows
- Host address
- AETitle = dcm4chee
- Port = 11112
- Q&R = Y
- Description = DCM4CHEE
- Transfer Syntax = Explicit Little Endian
- Configure the Client on the PACS server under “AE Management” using the settings from OSIRIX->Preferences->Listener Window
- Note: PACS Servers requires the DICOM IDs from the clients (as far as i know), and the AETitle match is important between the client and server.
Uploading DICOM images to your PACS server
The last step of the process is to be able to upload your DICOM images to the PACS server. In order to do this, a “Patient ID” is required or else the dcm4chee PACS server will not be able to accept the images.
- To insert a patient ID, ANONYMIZE the DICOM and insert your own patient ID by right clicking on the target DICOM. Once that is done, you will have saved a new set of images based on your defined ID.
- Delete the original DICOM images and import the new anonymized set of DICOM.
- Select the target DICOM and click “Send” in the icon bar
- Set the parameters of upload and send the data set to the PACS server.
At this stage, you can login to the PACS server and see that a new patient record would have appeared. Once the upload is complete, you would be clear to delete the DICOM from your client workstation and rely on the copy on server.
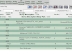
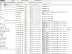
To download a set of DICOM images, you only need to QUERY the server and simply select the patient that you want to download by searching for the target. Once you have found the patient that you are looking for, you only need to click the download window to fetch the image into your computer. You can reference the screens you will see, in both OSIRIX and dcm4chee, in the images attached.
Some might be wondering how fast the uploads are. I have tested using a MacBook Pro client and a Mac Mini as a PACS server and transferred about 10GB worth of DICOM (9.77GB to be exact). The entire transfer took about 1/2 hour. I suppose this gives a pretty ok estimate on the expected waiting time on the uploading of DICOM images.
References
- ARRS Goldminers – ARRS GoldMiner provides rapid access to published, peer-reviewed medical images.
- Aycan – FDA approved version of OSIRIX
List of free DICOM images available:-
- Osirix Homepage
- http://idoimaging.com/cgi-bin/imaging/formats.pl?ident=2
- http://www.barre.nom.fr/medical/samples/
- http://www.mathworks.com/company/newsletters/digest/nov02/dicom.html
- http://www.mathworks.com/matlabcentral/fileexchange/loadFile.do?objectId=2762&objectType=file
- http://149.142.216.30/DICOM_FILES/DICOMfilesIndex.html
- http://www.wolfram.com/products/webmathematica/resources/dicom.htmlv
- Visible Human Project from the US National Library of Medicine
Commercial Tools
Some other commercial tools available:-
- HeartIT – WebPAX Medical Imaging Management System (http://www.heartit.com/index.html) – This is primarily a data store that allows users to access the WebPAX system over the Internet. It is more useful for being able to show movies of medical images over the Internet. It doesn’t really support extensive annotation and interactivity to those movies. There by limiting it to more of being a viewer.
- NeuroLens (http://www.neurolens.org/NeuroLens/Home.html) – This is an application that places DICOM and MRI data together for better analysis. Interestingly, this application is available free for Academic and non-profit research use.
- ResolutionMD by Calgary Scientific (http://www.calgaryscientific.com/calscimed/products.htm) – This tool is very much like OSIRIX, but apparently has a rather powerful “lens” tool that allows one to zoom up really close on the area of interest. It is also able to view in 2D and 3D with various coloring and rendering modes. Clipping is also possible. I haven’t encountered being able to annotate the image or view, but you can save a bookmark to a particular view.
- VG Studio Max by Volume Graphics (http://www.volumegraphics.com/products/vgstudiomax/index.html) – This is essentially a non-clinical modeling and rendering too mean to easily render DICOM easily and provide walk-through in a rather accurate fashion. This is in-effect a tool that is very good for education as it provides attractive visual output and views. it is also able to do slicing and clipping. In my opinion. VG Studio provides a good way to be able to take DICOM and output it into attractive videos for students to see and appreciate.